
Research on Protein-Membrane
About one fourth of the all proteins are membrane proteins, which are target of most of common drugs. They regulate cellular signaling, transport, and are part of our immune system and as such are related to number of deadly diseases ranging from cancer to Alzheimer’s disease. Yet our knowledge about them is very limited, e.g. membrane proteins represent only 4% of known structures in PDB database and we do not know how their function and distribution is related to specific membranes, i.e. how they are organized in space and time not even mention to specific organelles or cells. Using computer simulations we investigate the protein-membrane interactions at multi-scale level (from all-atom to very coarse grained) with focus on lipid specific effect. Such information, including our current results, can be used for the risks diagnostics, development new therapeutic peptides (antimicrobial peptides, sensors, target specific drug carriers), and suggestion of new treatment strategies including lipid diet and metabolism.
Most important projects are:
1. Antimicrobial peptides - More than 0.7 million deaths/year are caused by drug resistant bacteria and the number is estimated to grow to 10 million in 2050. This is why a growing microbial resistance together with slower development of new antibiotic was identified as serious threat to global public health by the World Health Organization, which encourages development of new treatments and drugs. Antimicrobial peptides are one of the candidates for such a new treatment. With the right properties, they can kill bacteria, viruses, or even cancer cells by disrupting their membranes and they are of the innate immune system of many organisms. By means of multi-scale computer simulations we systematically investigate the complex interplay between model peptides and the most common lipids from bacteria and human plasma membranes in order to identify crucial molecular features (length, flexibility, and distribution of hydrophobicity) that lead to translocation and pore formation at various membranes. The provided molecular understanding is useful for the rational design of new antimicrobial peptides used in medicine, cosmetics, or disinfection.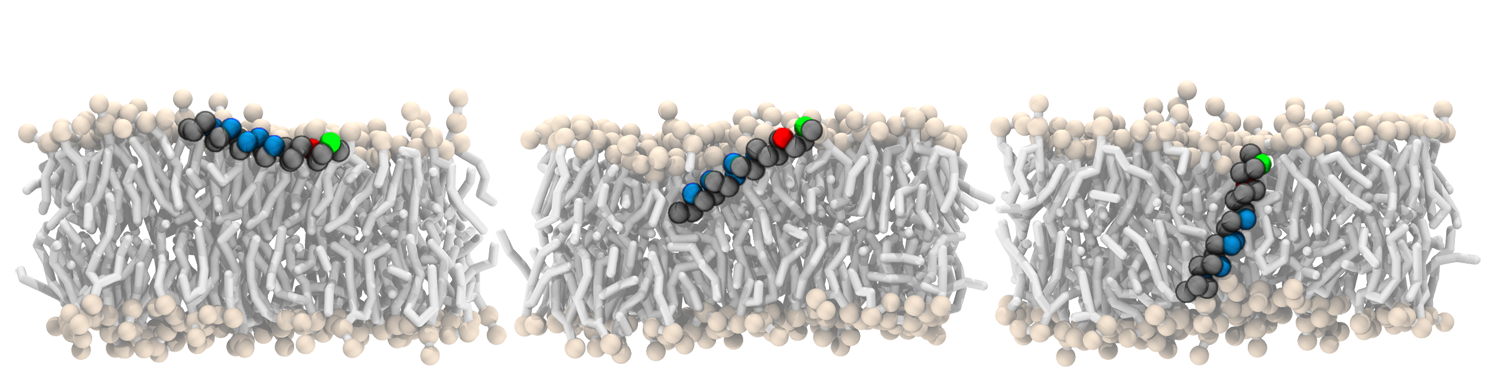
2. Signaling proteins - Proteins involved in signaling pathways are vital for cells and their malfunction is typically related to cancer or serious disease. We will focus on proteins that are associated with phospholipid membranes, which are crucial not only for protein proper function but also for correct distribution/positioning within the cell. We use computer simulations to study three specific proteins: [1] Dishavelled protein (DVL) from Wnt signaling pathway; [2] protein kinase D; and [3] DivIVA protein binding to negatively curved membranes, for which we have experimental collaborations.
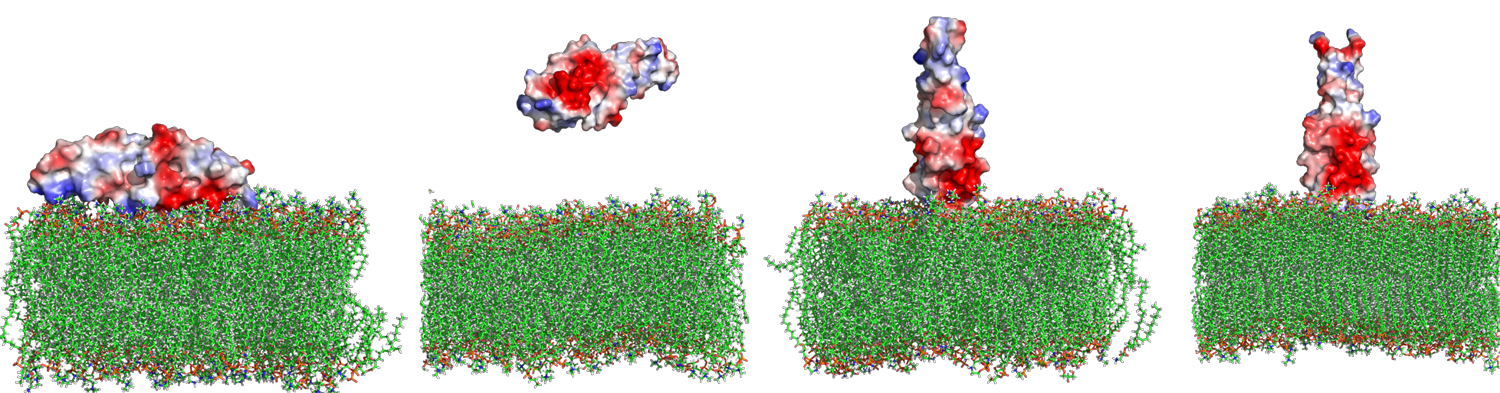
3. Abeta peptides - Alzheimer's Disease (AD) is the most common form of dementia in the elderly. Currently, 1 in 8 persons above the age of 65 is suffering from this disorder and it is expected that there will be 115 million persons affected globally by 2050 with treatment expenses over 2 billion$/year.[1] Amyloid-β (Aβ) peptide has been identified as a clinical hallmark in disease development and progression. Its oligomers are recognized as the most toxic disrupting neuronal membranes. However, the relation between the membrane composition and its effect on Aβ oligomerisation remains unclear. We have recently shown how Aβ oligomerisation can be prevented by physiological concentrations of one such ganglioside, GM1. Using state-of-the-art molecular dynamics simulation with advanced sampling techniques, coupled with single-molecule fluorescence microscopy and spectroscopy (see collaborations), we investigate other neuronal gangliosides in order to provide an atomic-level understanding of their interaction with Aβ peptides, which could be used in clinical applications.

