
Research on Protein-Protein Interactions
Proteins can aggregate in a wide variety of structures, which can be used to spontaneous formation of new peptide based materials or machines.
Most important projects are:
1. Virus Proteins – Viruses are ubiquitous deadly machines that cause wide range of diseases from common cold and flu to lethal HIV, Ebola, or SARS. Due to the recent progress in electron microscopy an increasing number of virus structures is being determined. The next step to fight the viruses is to determine the molecular mechanism of their action and find the ways to prevent it. We investigate the interactions and dynamics of proteins from viruses with main focus on structural changes related to infection and virus interaction with membrane. This is done in close collaboration with experiments including high-speed atomic force microscopy and cryo-electron microscopy. To this end we are part od Protein Dynamic Center.
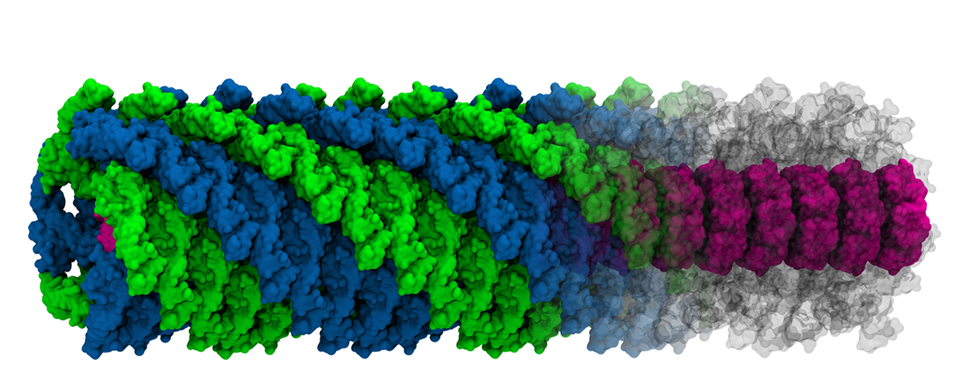
2. Peptide based materials and machines - Peptide- or protein-based materials have exceptional properties as evident from natural materials such as silk, hair, and bone. The capability to developing man-made materials with similar or enhanced characteristics would have an enormous impact on humankind. However, the ability to artificially create similar hierarchical materials is still stymied by multiple scientific and technical challenges. The major challenges in such a bottom-up design process are: [1] having a molecular understanding of the self-assembly of basic building blocks under different conditions, [2] control of the organisation of higher order structures that span large scales, [3] understanding the impact of the aggregate structures on living organisms, and [4] controlling the interaction with inorganic materials, e.g., mineralization. We focuse on amphiphilic peptides using top-down approach, where basic understanding is obtained from highly coarse- grained models and then further refined with a more detailed model. Multiple coarse-grained levels allow us to access large-scale changes and to obtain sequence specificity at the same time.


